This web page was produced as an assignment for Genetics 677, an undergraduate course at UW-Madison
DNA Motif Identification
Using the MOTIF Search I was unable to find any motif results using the DNA sequence of SMN1. A variety of parameters continued to yield a null result. However, using the mRNA sequence I was able to find a multitude of Motifs. Using a cut-off score of 95 I found 16 motifs and have presented the results of that search below.
| Motif Name | Number of Occurences | Description |
| HSF | 22 | Heat shock factor(drosophila) |
| HSF | 20 | Heat shock factor(yeast) |
| ADR1 | 5 | Alcohol dehydrogenase gene regulator 1 |
| Nkx-2.5 | 1 | Homeo domain factor Nkx-2.5/Csx, tinman homolog |
| AML-1a | 2 | Runt-factor AML-1 |
| c-Myb | 1 | c-Myb |
| CdxA | 1 | CdxA |
| Cap | 3 | Cap signal for transcription initiation |
| E4BP4 | 1 | E4BP4 |
| Skn-1 | 1 | Maternal gene product |
| GAmyb | 1 | GA-regulated myb gene from barley |
| GATA-1 | 1 | GATA-binding factor 1 |
| Oct-1 | 1 | Octamer factor 1 |
| TCF11 | 1 | TCF11/KCR-F1/Nrf1 homodimers |
| Dof2 | 1 | Dof2 – single zinc finger transcription factor |
| NIT2 | 2 | Activator of nitrogen-regulated genes |
MEME Motifs
Using the MEME program I was able to identify 3 motifs on the original DNA sequence of SMN1.
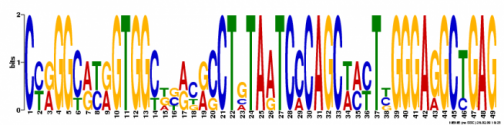
Motif 1
Width: 49 nucleotides
Occurence: 5 times
Likelihood Ratio: 275
E-Value: 8.4e-026
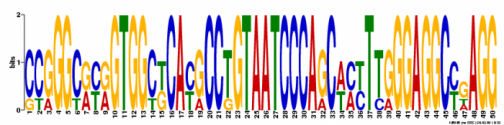
Motif 2
Width: 50 nucleotides
Occurence: 5 times
Likelihood Ratio: 300
E-Value: 4.1e-034
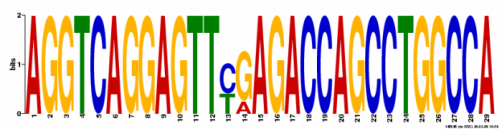
Motif 3
Width: 29 nucleotides
Occurence: 5 times
Likelihood Ratio: 201
E-Value: 7.0e-023
Analysis
The motifs that were discovered using the MEME program were all from the DNA gene sequence. It is strange to see that there were no motifs found using the Motif search program. I would expect the results of both programs to be reasonably similar. It seems that they used different methods of obtaining motif information which could result in these differences. The MEME program searched the sequence for similarities in DNA patterns within the DNA of SMN1. The Motif search examined existing databases for motifs that have already been identified. The difference in the search will lead to differences, but no similarity at all is very strange, especially considering that no DNA motifs were recognized by Motif Search when the original DNA sequence was tested.
Motif Search was very easy to use and the information was presented in an easy to understand way. The results were also obtained fairly quickly. The MEME program required a rather lengthy wait. It presented a lot of information but it seemed to be of a more technical sort. It was not the clear cut information that I was seeking, but it was still informative.
William Baader
[email protected]
Feb 26 2009
www.gen677.weebly.com